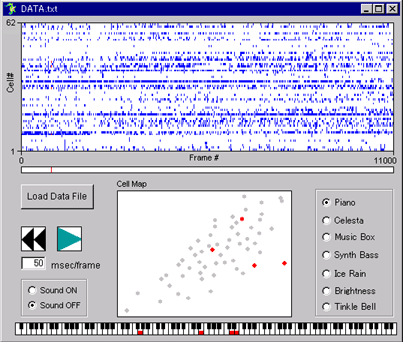
raw movie |
.gif) ΔF/F movie |
|
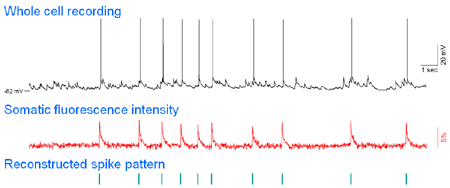 |
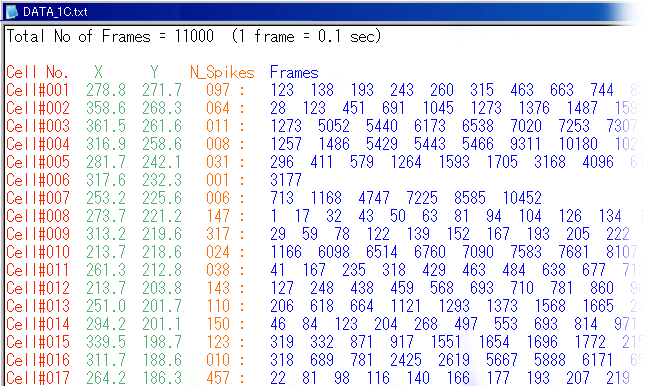
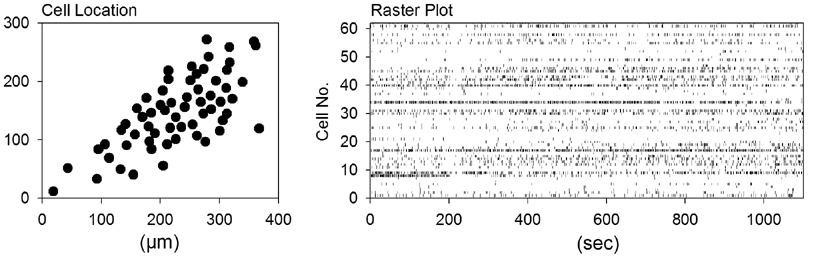
| data # | movie length | frame rate | number of neurons | region | temperature | |
|---|---|---|---|---|---|---|
| DATA_001.txt | 1100 sec | 10 Hz | 62 | hippocampal CA3 | 32 ºC | |
| DATA_002.txt | 700 sec | 10 Hz | 126 | hippocampal CA3 | 32 ºC | |
| DATA_003.txt | 600 sec | 10 Hz | 226 | hippocampal CA3 | 32 ºC | |
| DATA_004.txt | 1200 sec | 10 Hz | 64 | hippocampal CA3 | 32 ºC | |
| DATA_005.txt | 610 sec | 10 Hz | 88 | hippocampal CA3 | 32 ºC | |
| DATA_006.txt | 1200 sec | 10 Hz | 95 | hippocampal CA3 | 32 ºC | |
| DATA_007.txt | 310 sec | 10 Hz | 68 | hippocampal CA3 | 32 ºC | |
| DATA_008.txt | 1200 sec | 10 Hz | 93 | hippocampal CA3 | 32 ºC | |
temperature dependence |
||||||
| DATA_009.txt | 180 sec | 100 Hz | 169 | hippocampal CA3 | 24 ºC | |
| DATA_010.txt | 180 sec | 100 Hz | 106 | hippocampal CA3 | 24 ºC | |
| DATA_011.txt | 180 sec | 100 Hz | 135 | hippocampal CA3 | 24 ºC | |
| DATA_012.txt | 180 sec | 100 Hz | 121 | hippocampal CA3 | 24 ºC | |
| DATA_013.txt | 180 sec | 100 Hz | 120 | hippocampal CA3 | 24 ºC | |
| DATA_014.txt | 180 sec | 100 Hz | 156 | hippocampal CA3 | 28 ºC | |
| DATA_015.txt | 180 sec | 100 Hz | 105 | hippocampal CA3 | 28 ºC | |
| DATA_016.txt | 180 sec | 100 Hz | 134 | hippocampal CA3 | 28 ºC | |
| DATA_017.txt | 180 sec | 100 Hz | 112 | hippocampal CA3 | 28 ºC | |
| DATA_018.txt | 180 sec | 100 Hz | 134 | hippocampal CA3 | 28 ºC | |
| DATA_019.txt | 180 sec | 100 Hz | 127 | hippocampal CA3 | 32 ºC | |
| DATA_020.txt | 180 sec | 100 Hz | 179 | hippocampal CA3 | 32 ºC | |
| DATA_021.txt | 180 sec | 100 Hz | 150 | hippocampal CA3 | 32 ºC | |
| DATA_022.txt | 180 sec | 100 Hz | 125 | hippocampal CA3 | 32 ºC | |
| DATA_023.txt | 180 sec | 100 Hz | 135 | hippocampal CA3 | 32 ºC | |
| DATA_024.txt | 180 sec | 100 Hz | 105 | hippocampal CA3 | 36 ºC | |
| DATA_025.txt | 180 sec | 100 Hz | 157 | hippocampal CA3 | 36 ºC | |
| DATA_026.txt | 180 sec | 100 Hz | 137 | hippocampal CA3 | 36 ºC | |
| DATA_027.txt | 180 sec | 100 Hz | 120 | hippocampal CA3 | 36 ºC | |
| DATA_028.txt | 180 sec | 100 Hz | 152 | hippocampal CA3 | 36 ºC | |
| DATA_029.txt | 180 sec | 100 Hz | 129 | hippocampal CA3 | 40 ºC | |
| DATA_030.txt | 180 sec | 100 Hz | 106 | hippocampal CA3 | 40 ºC | |
| DATA_031.txt | 180 sec | 100 Hz | 187 | hippocampal CA3 | 40 ºC | |
| DATA_032.txt | 180 sec | 100 Hz | 160 | hippocampal CA3 | 40 ºC | |
| DATA_033.txt | 180 sec | 100 Hz | 161 | hippocampal CA3 | 40 ºC | |
| DATA_034.txt | 180 sec | 200 Hz | 74 | hippocampal CA3 | 32 ºC | |
fast scanning |
||||||
| DATA_035.txt | 130 sec | 500 Hz | 60 | hippocampal CA3 | 32 ºC | |
| DATA_036.txt | 130 sec | 500 Hz | 88 | hippocampal CA3 | 32 ºC | |
| DATA_037.txt | 130 sec | 500 Hz | 88 | hippocampal CA3 | 32 ºC | |
| DATA_038.txt | 130 sec | 500 Hz | 64 | hippocampal CA3 | 32 ºC | |
| DATA_039.txt | 130 sec | 500 Hz | 96 | hippocampal CA3 | 32 ºC | |
| DATA_040.txt | 130 sec | 500 Hz | 137 | hippocampal CA3 | 32 ºC | |
| DATA_041.txt | 130 sec | 500 Hz | 87 | hippocampal CA3 | 32 ºC | |
| DATA_042.txt | 130 sec | 500 Hz | 95 | hippocampal CA3 | 32 ºC | |
| DATA_043.txt | 130 sec | 500 Hz | 85 | hippocampal CA3 | 32 ºC | |
| DATA_044.txt | 130 sec | 500 Hz | 131 | hippocampal CA3 | 32 ºC | |
| DATA_045.txt | 130 sec | 500 Hz | 77 | hippocampal CA3 | 32 ºC | |
| DATA_046.txt | 130 sec | 500 Hz | 53 | hippocampal CA3 | 32 ºC | |
| DATA_047.txt | 130 sec | 500 Hz | 72 | hippocampal CA3 | 32 ºC | |
| DATA_048.txt | 130 sec | 500 Hz | 60 | hippocampal CA3 | 32 ºC | |
long-time movie |
||||||
| DATA_049.txt | 3300 sec | 10 Hz | 140 | hippocampal CA3 | 32 ºC | |